Note
Click here to download the full example code
PyTorch Profiler¶
This recipe explains how to use PyTorch profiler and measure the time and memory consumption of the model’s operators.
Introduction¶
PyTorch includes a simple profiler API that is useful when user needs to determine the most expensive operators in the model.
In this recipe, we will use a simple Resnet model to demonstrate how to use profiler to analyze model performance.
Steps¶
Import all necessary libraries
Instantiate a simple Resnet model
Using profiler to analyze execution time
Using profiler to analyze memory consumption
Using tracing functionality
Examining stack traces
Using profiler to analyze long-running jobs
1. Import all necessary libraries¶
In this recipe we will use torch, torchvision.models
and profiler modules:
import torch
import torchvision.models as models
from torch.profiler import profile, record_function, ProfilerActivity
2. Instantiate a simple Resnet model¶
Let’s create an instance of a Resnet model and prepare an input for it:
model = models.resnet18()
inputs = torch.randn(5, 3, 224, 224)
3. Using profiler to analyze execution time¶
PyTorch profiler is enabled through the context manager and accepts a number of parameters, some of the most useful are:
activities- a list of activities to profile:ProfilerActivity.CPU- PyTorch operators, TorchScript functions and user-defined code labels (seerecord_functionbelow);ProfilerActivity.CUDA- on-device CUDA kernels;
record_shapes- whether to record shapes of the operator inputs;profile_memory- whether to report amount of memory consumed by model’s Tensors;use_cuda- whether to measure execution time of CUDA kernels.
Note: when using CUDA, profiler also shows the runtime CUDA events occurring on the host.
Let’s see how we can use profiler to analyze the execution time:
with profile(activities=[ProfilerActivity.CPU], record_shapes=True) as prof:
with record_function("model_inference"):
model(inputs)
Note that we can use record_function context manager to label
arbitrary code ranges with user provided names
(model_inference is used as a label in the example above).
Profiler allows one to check which operators were called during the
execution of a code range wrapped with a profiler context manager.
If multiple profiler ranges are active at the same time (e.g. in
parallel PyTorch threads), each profiling context manager tracks only
the operators of its corresponding range.
Profiler also automatically profiles the asynchronous tasks launched
with torch.jit._fork and (in case of a backward pass)
the backward pass operators launched with backward() call.
Let’s print out the stats for the execution above:
print(prof.key_averages().table(sort_by="cpu_time_total", row_limit=10))
The output will look like (omitting some columns):
# --------------------------------- ------------ ------------ ------------ ------------
# Name Self CPU CPU total CPU time avg # of Calls
# --------------------------------- ------------ ------------ ------------ ------------
# model_inference 5.509ms 57.503ms 57.503ms 1
# aten::conv2d 231.000us 31.931ms 1.597ms 20
# aten::convolution 250.000us 31.700ms 1.585ms 20
# aten::_convolution 336.000us 31.450ms 1.573ms 20
# aten::mkldnn_convolution 30.838ms 31.114ms 1.556ms 20
# aten::batch_norm 211.000us 14.693ms 734.650us 20
# aten::_batch_norm_impl_index 319.000us 14.482ms 724.100us 20
# aten::native_batch_norm 9.229ms 14.109ms 705.450us 20
# aten::mean 332.000us 2.631ms 125.286us 21
# aten::select 1.668ms 2.292ms 8.988us 255
# --------------------------------- ------------ ------------ ------------ ------------
# Self CPU time total: 57.549m
#
Here we see that, as expected, most of the time is spent in convolution (and specifically in mkldnn_convolution
for PyTorch compiled with MKL-DNN support).
Note the difference between self cpu time and cpu time - operators can call other operators, self cpu time excludes time
spent in children operator calls, while total cpu time includes it. You can choose to sort by the self cpu time by passing
sort_by="self_cpu_time_total" into the table call.
To get a finer granularity of results and include operator input shapes, pass group_by_input_shape=True
(note: this requires running the profiler with record_shapes=True):
print(prof.key_averages(group_by_input_shape=True).table(sort_by="cpu_time_total", row_limit=10))
The output might look like this (omitting some columns):
--------------------------------- ------------ -------------------------------------------
Name CPU total Input Shapes
--------------------------------- ------------ -------------------------------------------
model_inference 57.503ms []
aten::conv2d 8.008ms [5,64,56,56], [64,64,3,3], [], ..., []]
aten::convolution 7.956ms [[5,64,56,56], [64,64,3,3], [], ..., []]
aten::_convolution 7.909ms [[5,64,56,56], [64,64,3,3], [], ..., []]
aten::mkldnn_convolution 7.834ms [[5,64,56,56], [64,64,3,3], [], ..., []]
aten::conv2d 6.332ms [[5,512,7,7], [512,512,3,3], [], ..., []]
aten::convolution 6.303ms [[5,512,7,7], [512,512,3,3], [], ..., []]
aten::_convolution 6.273ms [[5,512,7,7], [512,512,3,3], [], ..., []]
aten::mkldnn_convolution 6.233ms [[5,512,7,7], [512,512,3,3], [], ..., []]
aten::conv2d 4.751ms [[5,256,14,14], [256,256,3,3], [], ..., []]
--------------------------------- ------------ -------------------------------------------
Self CPU time total: 57.549ms
Note the occurrence of aten::convolution twice with different input shapes.
Profiler can also be used to analyze performance of models executed on GPUs:
model = models.resnet18().cuda()
inputs = torch.randn(5, 3, 224, 224).cuda()
with profile(activities=[
ProfilerActivity.CPU, ProfilerActivity.CUDA], record_shapes=True) as prof:
with record_function("model_inference"):
model(inputs)
print(prof.key_averages().table(sort_by="cuda_time_total", row_limit=10))
(Note: the first use of CUDA profiling may bring an extra overhead.)
The resulting table output (omitting some columns):
------------------------------------------------------- ------------ ------------
Name Self CUDA CUDA total
------------------------------------------------------- ------------ ------------
model_inference 0.000us 11.666ms
aten::conv2d 0.000us 10.484ms
aten::convolution 0.000us 10.484ms
aten::_convolution 0.000us 10.484ms
aten::_convolution_nogroup 0.000us 10.484ms
aten::thnn_conv2d 0.000us 10.484ms
aten::thnn_conv2d_forward 10.484ms 10.484ms
void at::native::im2col_kernel<float>(long, float co... 3.844ms 3.844ms
sgemm_32x32x32_NN 3.206ms 3.206ms
sgemm_32x32x32_NN_vec 3.093ms 3.093ms
------------------------------------------------------- ------------ ------------
Self CPU time total: 23.015ms
Self CUDA time total: 11.666ms
Note the occurrence of on-device kernels in the output (e.g. sgemm_32x32x32_NN).
4. Using profiler to analyze memory consumption¶
PyTorch profiler can also show the amount of memory (used by the model’s tensors)
that was allocated (or released) during the execution of the model’s operators.
In the output below, ‘self’ memory corresponds to the memory allocated (released)
by the operator, excluding the children calls to the other operators.
To enable memory profiling functionality pass profile_memory=True.
model = models.resnet18()
inputs = torch.randn(5, 3, 224, 224)
with profile(activities=[ProfilerActivity.CPU],
profile_memory=True, record_shapes=True) as prof:
model(inputs)
print(prof.key_averages().table(sort_by="self_cpu_memory_usage", row_limit=10))
# (omitting some columns)
# --------------------------------- ------------ ------------ ------------
# Name CPU Mem Self CPU Mem # of Calls
# --------------------------------- ------------ ------------ ------------
# aten::empty 94.79 Mb 94.79 Mb 121
# aten::max_pool2d_with_indices 11.48 Mb 11.48 Mb 1
# aten::addmm 19.53 Kb 19.53 Kb 1
# aten::empty_strided 572 b 572 b 25
# aten::resize_ 240 b 240 b 6
# aten::abs 480 b 240 b 4
# aten::add 160 b 160 b 20
# aten::masked_select 120 b 112 b 1
# aten::ne 122 b 53 b 6
# aten::eq 60 b 30 b 2
# --------------------------------- ------------ ------------ ------------
# Self CPU time total: 53.064ms
print(prof.key_averages().table(sort_by="cpu_memory_usage", row_limit=10))
The output might look like this (omitting some columns):
--------------------------------- ------------ ------------ ------------
Name CPU Mem Self CPU Mem # of Calls
--------------------------------- ------------ ------------ ------------
aten::empty 94.79 Mb 94.79 Mb 121
aten::batch_norm 47.41 Mb 0 b 20
aten::_batch_norm_impl_index 47.41 Mb 0 b 20
aten::native_batch_norm 47.41 Mb 0 b 20
aten::conv2d 47.37 Mb 0 b 20
aten::convolution 47.37 Mb 0 b 20
aten::_convolution 47.37 Mb 0 b 20
aten::mkldnn_convolution 47.37 Mb 0 b 20
aten::max_pool2d 11.48 Mb 0 b 1
aten::max_pool2d_with_indices 11.48 Mb 11.48 Mb 1
--------------------------------- ------------ ------------ ------------
Self CPU time total: 53.064ms
5. Using tracing functionality¶
Profiling results can be outputted as a .json trace file:
model = models.resnet18().cuda()
inputs = torch.randn(5, 3, 224, 224).cuda()
with profile(activities=[ProfilerActivity.CPU, ProfilerActivity.CUDA]) as prof:
model(inputs)
prof.export_chrome_trace("trace.json")
You can examine the sequence of profiled operators and CUDA kernels
in Chrome trace viewer (chrome://tracing):
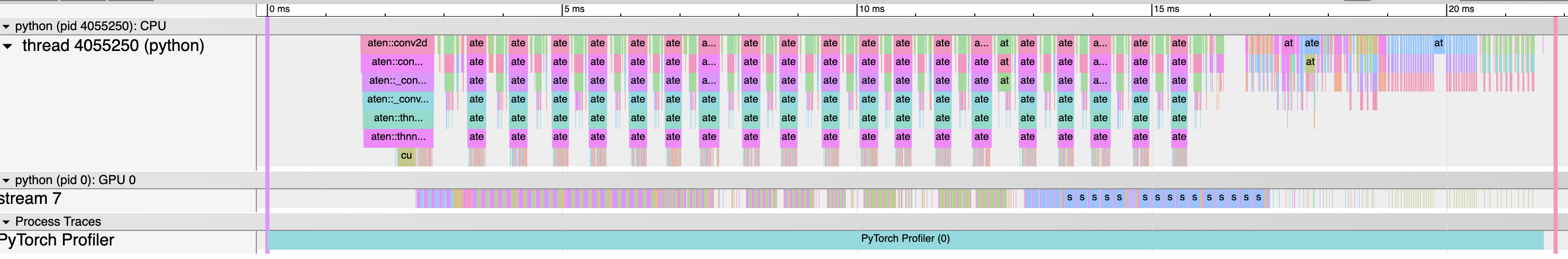
6. Examining stack traces¶
Profiler can be used to analyze Python and TorchScript stack traces:
with profile(
activities=[ProfilerActivity.CPU, ProfilerActivity.CUDA],
with_stack=True,
) as prof:
model(inputs)
# Print aggregated stats
print(prof.key_averages(group_by_stack_n=5).table(sort_by="self_cuda_time_total", row_limit=2))
The output might look like this (omitting some columns):
------------------------- -----------------------------------------------------------
Name Source Location
------------------------- -----------------------------------------------------------
aten::thnn_conv2d_forward .../torch/nn/modules/conv.py(439): _conv_forward
.../torch/nn/modules/conv.py(443): forward
.../torch/nn/modules/module.py(1051): _call_impl
.../site-packages/torchvision/models/resnet.py(63): forward
.../torch/nn/modules/module.py(1051): _call_impl
aten::thnn_conv2d_forward .../torch/nn/modules/conv.py(439): _conv_forward
.../torch/nn/modules/conv.py(443): forward
.../torch/nn/modules/module.py(1051): _call_impl
.../site-packages/torchvision/models/resnet.py(59): forward
.../torch/nn/modules/module.py(1051): _call_impl
------------------------- -----------------------------------------------------------
Self CPU time total: 34.016ms
Self CUDA time total: 11.659ms
Note the two convolutions and the two call sites in torchvision/models/resnet.py script.
(Warning: stack tracing adds an extra profiling overhead.)
7. Using profiler to analyze long-running jobs¶
PyTorch profiler offers an additional API to handle long-running jobs (such as training loops). Tracing all of the execution can be slow and result in very large trace files. To avoid this, use optional arguments:
schedule- specifies a function that takes an integer argument (step number) as an input and returns an action for the profiler, the best way to use this parameter is to usetorch.profiler.schedulehelper function that can generate a schedule for you;on_trace_ready- specifies a function that takes a reference to the profiler as an input and is called by the profiler each time the new trace is ready.
To illustrate how the API works, let’s first consider the following example with
torch.profiler.schedule helper function:
Profiler assumes that the long-running job is composed of steps, numbered starting from zero. The example above defines the following sequence of actions for the profiler:
Parameter
skip_firsttells profiler that it should ignore the first 10 steps (default value ofskip_firstis zero);After the first
skip_firststeps, profiler starts executing profiler cycles;Each cycle consists of three phases:
idling (
wait=5steps), during this phase profiler is not active;warming up (
warmup=1steps), during this phase profiler starts tracing, but the results are discarded; this phase is used to discard the samples obtained by the profiler at the beginning of the trace since they are usually skewed by an extra overhead;active tracing (
active=3steps), during this phase profiler traces and records data;
An optional
repeatparameter specifies an upper bound on the number of cycles. By default (zero value), profiler will execute cycles as long as the job runs.
Thus, in the example above, profiler will skip the first 15 steps, spend the next step on the warm up,
actively record the next 3 steps, skip another 5 steps, spend the next step on the warm up, actively
record another 3 steps. Since the repeat=2 parameter value is specified, the profiler will stop
the recording after the first two cycles.
At the end of each cycle profiler calls the specified on_trace_ready function and passes itself as
an argument. This function is used to process the new trace - either by obtaining the table output or
by saving the output on disk as a trace file.
To send the signal to the profiler that the next step has started, call prof.step() function.
The current profiler step is stored in prof.step_num.
The following example shows how to use all of the concepts above:
def trace_handler(p):
output = p.key_averages().table(sort_by="self_cuda_time_total", row_limit=10)
print(output)
p.export_chrome_trace("/tmp/trace_" + str(p.step_num) + ".json")
with profile(
activities=[ProfilerActivity.CPU, ProfilerActivity.CUDA],
schedule=torch.profiler.schedule(
wait=1,
warmup=1,
active=2),
on_trace_ready=trace_handler
) as p:
for idx in range(8):
model(inputs)
p.step()
Learn More¶
Take a look at the following recipes/tutorials to continue your learning:
PyTorch Profiler with TensorBoard tutorial
Visualizing models, data, and training with TensorBoard tutorial
Total running time of the script: ( 0 minutes 0.000 seconds)